The primary emphasis of the Meyers lab is the analysis of small RNAs in plants. With our many collaborators, we have pioneered genomic analysis of small RNAs and their targets, working with “next-gen” sequencing technologies nearly since their invention.
Our work with next-gen sequencing stretches back to ~2001, when Blake was funded by the NSF to apply “MPSS” to the analysis of gene expression in Arabidopsis. This led to the development in the Meyers lab of the first publicly-accessible browser for next-gen data.
After moving to the University of Delaware in 2002, we continued to develop the application to mRNA and small RNA analyses of first MPSS, then 454, and finally the still-current Illumina SBS sequencing.
In 2005, with collaborator Pam Green, we were the first to use next-gen sequencing for the analysis of small RNAs, and in 2008, our labs co-developed “PARE” for the genome-wide analysis of cleaved mRNAs (PARE = Parallel Analysis of RNA Ends).
The Meyers lab has widely applied these methods to study plant genomes and their RNA products, and the lab continues to develop and apply novel informatics approaches for the analysis of RNA function in plants.
Specific areas of research include the use of these technologies to assess small RNA function and biogenesis in a broad range of plants, including Arabidopsis, maize, soybean, rice, and diverse other species. These data are being used to identify novel transcripts, to study small RNAs, microRNA targets, alternatively-polyadenylated transcripts, non-coding RNAs and gene silencing. The data are being analyzed to determine patterns of gene expression under different developmental conditions, for example to identify tissue-specific gene expression.


In 2005, with collaborator Pam Green, we were the first to use next-gen sequencing for the analysis of small RNAs, and in 2008, our labs co-developed “PARE” for the genome-wide analysis of cleaved mRNAs (PARE = Parallel Analysis of RNA Ends).
The Meyers lab has widely applied these methods to study plant genomes and their RNA products, and the lab continues to develop and apply novel informatics approaches for the analysis of RNA function in plants.
Specific areas of research include the use of these technologies to assess small RNA function and biogenesis in a broad range of plants, including Arabidopsis, maize, soybean, rice, and diverse other species. These data are being used to identify novel transcripts, to study small RNAs, microRNA targets, alternatively-polyadenylated transcripts, non-coding RNAs and gene silencing. The data are being analyzed to determine patterns of gene expression under different developmental conditions, for example to identify tissue-specific gene expression.
These studies are centered on the use of novel genomics tools and bioinformatics approaches to address fundamental questions of small RNA biology, gene expression, and genome regulation in plants.
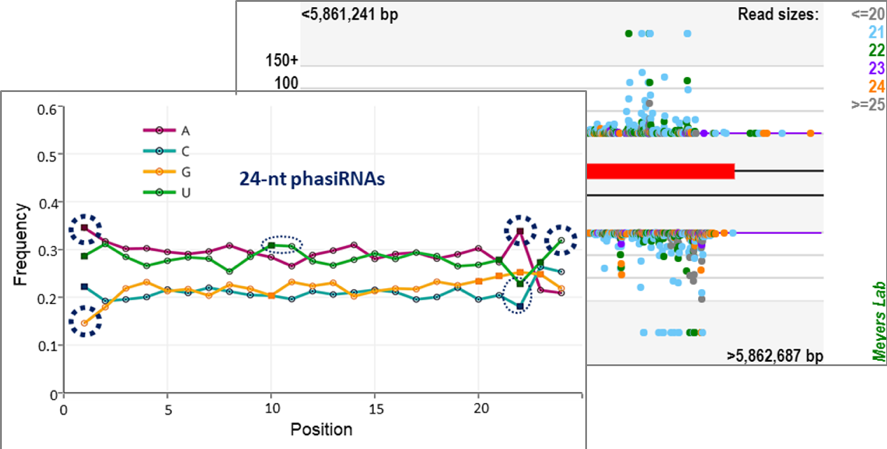
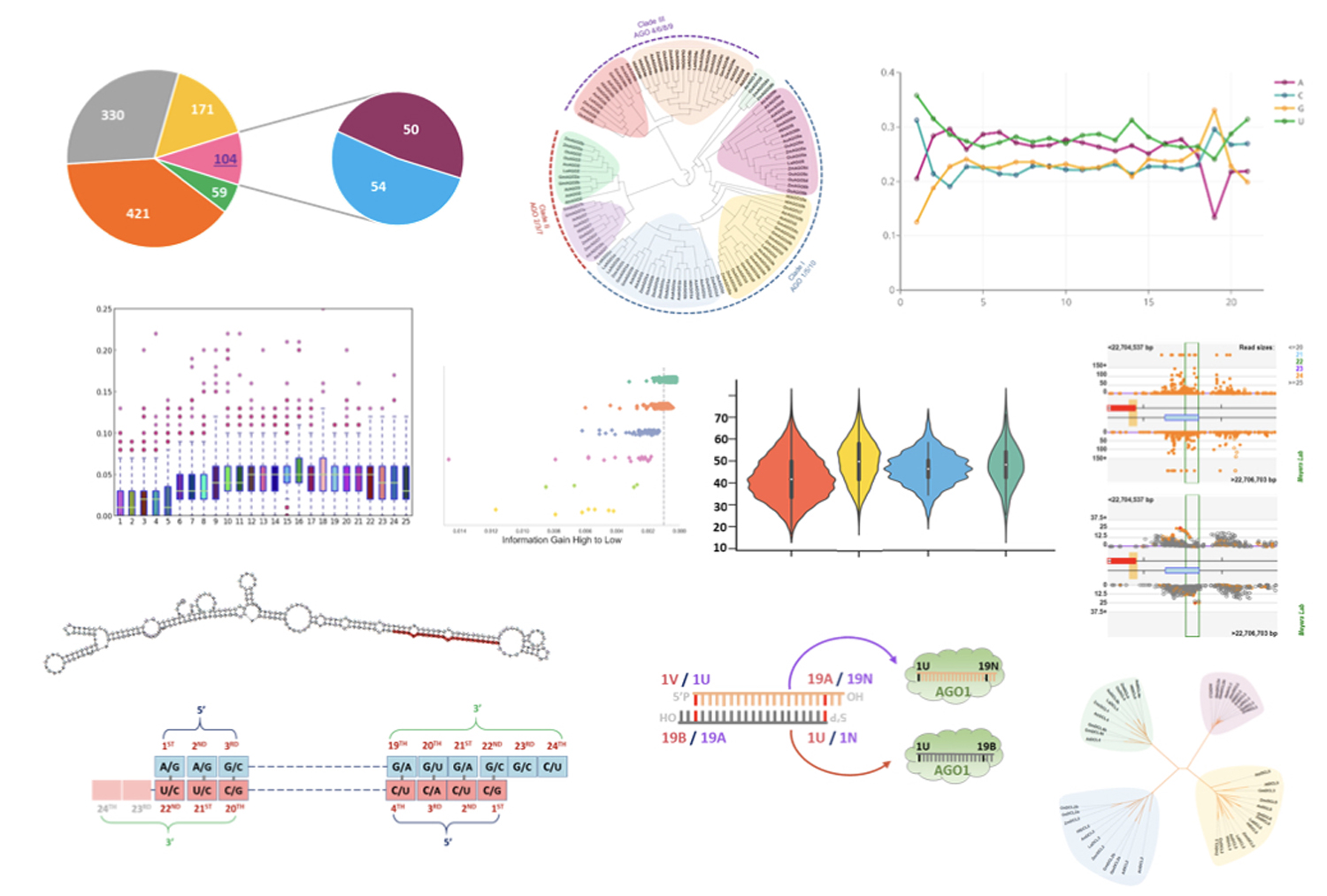
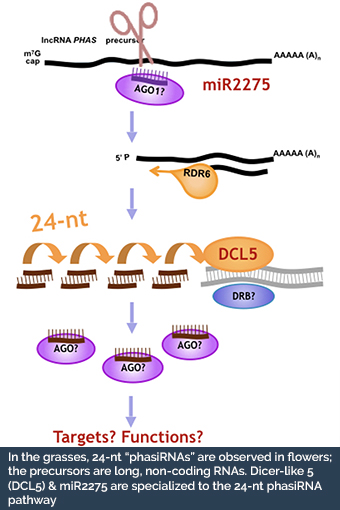
Phased, secondary, small interfering RNAs (phasiRNAs) are of particular interest to us. Originally designated as trans-acting small interfering RNAs or tasiRNAs, the wider group of phasiRNAs are triggered by microRNAs and produced as siRNAs. Like microRNAs, phasiRNA function in the suppression of target transcript levels.
In some plants, NB-LRRs have a particularly high level of redundancy in miRNA and phasiRNA-mediated regulation.
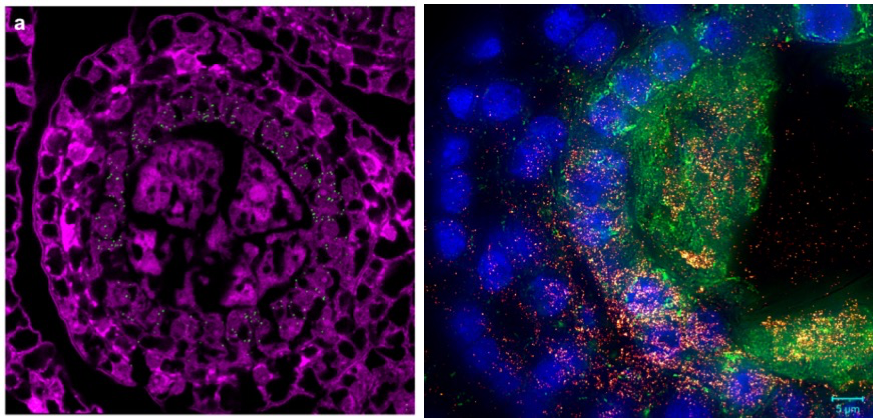
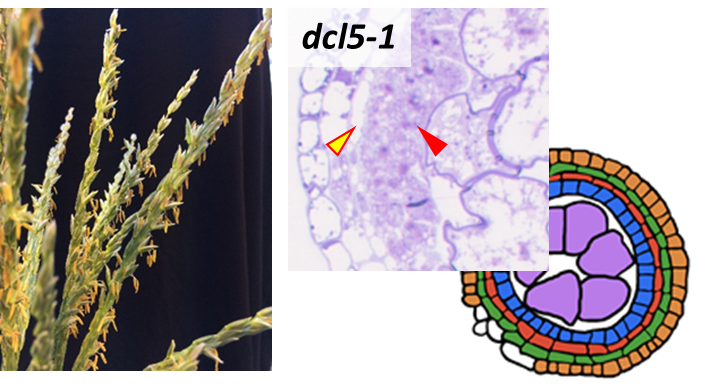
In the grasses, phasiRNAs from non-coding RNAs are prevalent in anthers, during early development and meiosis.
Finally, we retain a bit of a historical interest in disease resistance genes in plants. The Nucleotide Binding Site-Leucine Rich Repeat (NB-LRR) proteins encoded by many resistance genes provide the first line of defense in many specific plant-pathogen interactions.
Approximately 150 of these proteins are encoded in the Arabidopsis Col-0 genome; with variable numbers ranging up to hundreds per plant genome in other species. We study sequence variation, function, and evolution in this class of genes. Connecting two of our long-standing interests, we were the first to describe microRNAs as “master regulators” via direct and indirect targeting (phasiRNAs) of this gene family.


Genome Center
UC Davis

UC Davis
Genome Center